news model
license
































































































| ~ | 11696 (G/A) | 11696 (G/C) | 11696 (G/T) |
|---|---|---|---|
| ~ | 11696 (GTC/ATC) | 11696 (GTC/CTC) | 11696 (GTC/TTC) |
| MitImpact id | MI.18291 | MI.18292 | MI.18293 |
| Chr | chrM | chrM | chrM |
| Start | 11696 | 11696 | 11696 |
| Ref | G | G | G |
| Alt | A | C | T |
| Gene symbol | MT-ND4 | MT-ND4 | MT-ND4 |
| Extended annotation | mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 4 | mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 4 | mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 4 |
| Gene position | 937 | 937 | 937 |
| Gene start | 10760 | 10760 | 10760 |
| Gene end | 12137 | 12137 | 12137 |
| Gene strand | + | + | + |
| Codon substitution | GTC/ATC | GTC/CTC | GTC/TTC |
| AA position | 313 | 313 | 313 |
| AA ref | V | V | V |
| AA alt | I | L | F |
| Functional effect general | missense | missense | missense |
| Functional effect detailed | missense | missense | missense |
| OMIM id | 516003 | 516003 | 516003 |
| HGVS | NC_012920.1:g.11696G>A | NC_012920.1:g.11696G>C | NC_012920.1:g.11696G>T |
| HGNC id | 7459 | 7459 | 7459 |
| Respiratory Chain complex | I | I | I |
| Ensembl gene id | ENSG00000198886 | ENSG00000198886 | ENSG00000198886 |
| Ensembl transcript id | ENST00000361381 | ENST00000361381 | ENST00000361381 |
| Ensembl protein id | ENSP00000354961 | ENSP00000354961 | ENSP00000354961 |
| Uniprot id | P03905 | P03905 | P03905 |
| Uniprot name | NU4M_HUMAN | NU4M_HUMAN | NU4M_HUMAN |
| Ncbi gene id | 4538 | 4538 | 4538 |
| Ncbi protein id | YP_003024035.1 | YP_003024035.1 | YP_003024035.1 |
| PhyloP 100V | 1.117 | 1.117 | 1.117 |
| PhyloP 470Way | -0.26 | -0.26 | -0.26 |
| PhastCons 100V | 0.005 | 0.005 | 0.005 |
| PhastCons 470Way | 0.004 | 0.004 | 0.004 |
| PolyPhen2 | benign | benign | benign |
| PolyPhen2 score | 0.01 | 0.05 | 0.38 |
| SIFT | neutral | neutral | neutral |
| SIFT score | 0.7 | 1.0 | 0.11 |
| SIFT4G | Tolerated | Tolerated | Tolerated |
| SIFT4G score | 1.0 | 0.937 | 0.107 |
| VEST | Neutral | Neutral | Pathogenic |
| VEST pvalue | 0.28 | 0.15 | 0.04 |
| VEST FDR | 0.45 | 0.4 | 0.35 |
| Mitoclass.1 | neutral | neutral | neutral |
| SNPDryad | Neutral | Neutral | Neutral |
| SNPDryad score | 0.78 | 0.5 | 0.38 |
| MutationTaster | Disease automatic | Polymorphism | Polymorphism |
| MutationTaster score | 4.98847e-06 | 0.999993 | 1.0 |
| MutationTaster converted rankscore | 0.08975 | 0.08975 | 0.08975 |
| MutationTaster model | complex_aae | complex_aae | complex_aae |
| MutationTaster AAE | V313I | V313L | V313F |
| fathmm | Tolerated | Tolerated | Tolerated |
| fathmm score | 4.76 | 4.75 | 4.66 |
| fathmm converted rankscore | 0.01576 | 0.01589 | 0.01756 |
| AlphaMissense | likely_benign | likely_benign | likely_benign |
| AlphaMissense score | 0.0757 | 0.1458 | 0.2637 |
| CADD | Neutral | Neutral | Neutral |
| CADD score | -0.960482 | -0.814922 | 0.959151 |
| CADD phred | 0.019 | 0.04 | 10.43 |
| PROVEAN | Tolerated | Tolerated | Tolerated |
| PROVEAN score | -0.32 | -0.54 | -1.91 |
| MutationAssessor | neutral | neutral | neutral |
| MutationAssessor score | -1.085 | -1.485 | 0.8 |
| EFIN SP | Damaging | Neutral | Neutral |
| EFIN SP score | 0.284 | 0.702 | 0.73 |
| EFIN HD | Neutral | Neutral | Neutral |
| EFIN HD score | 0.338 | 0.912 | 0.592 |
| MLC | Deleterious | Deleterious | Deleterious |
| MLC score | 0.79349991 | 0.79349991 | 0.79349991 |
| PANTHER score | 0.333 | . | . |
| PhD-SNP score | 0.039 | . | . |
| APOGEE1 | Pathogenic | Neutral | Neutral |
| APOGEE1 score | 0.82 | 0.33 | 0.44 |
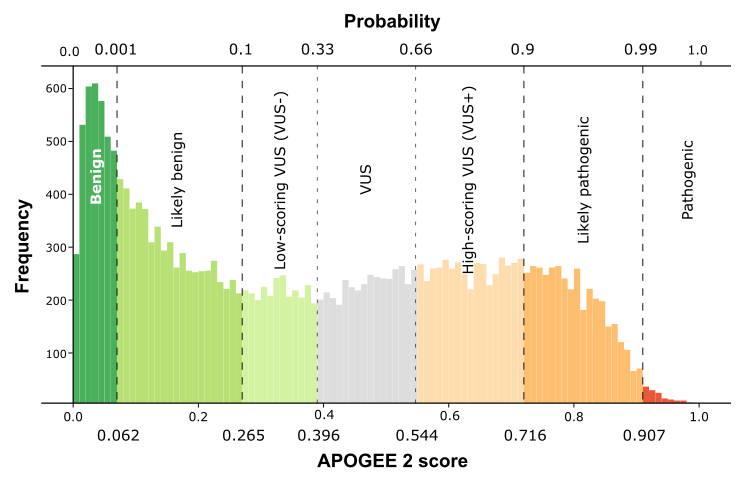
| APOGEE2 | VUS- | Likely-benign | VUS- |
| APOGEE2 score | 0.338351889297234 | 0.0712247635222073 | 0.289499771404907 |
| CAROL | neutral | neutral | neutral |
| CAROL score | 0.29 | 0.05 | 0.87 |
| Condel | deleterious | deleterious | neutral |
| Condel score | 0.85 | 0.98 | 0.37 |
| COVEC WMV | neutral | neutral | neutral |
| COVEC WMV score | -6 | -6 | -6 |
| MtoolBox | neutral | neutral | neutral |
| MtoolBox DS | 0.12 | 0.15 | 0.29 |
| DEOGEN2 | Tolerated | Tolerated | Tolerated |
| DEOGEN2 score | 0.013156 | 0.014771 | 0.031141 |
| DEOGEN2 converted rankscore | 0.11431 | 0.12515 | 0.21906 |
| Meta-SNP | Neutral | . | . |
| Meta-SNP score | 0.054 | . | . |
| PolyPhen2 transf | medium impact | medium impact | medium impact |
| PolyPhen2 transf score | 1.16 | 0.48 | -0.52 |
| SIFT_transf | medium impact | high impact | medium impact |
| SIFT transf score | 0.41 | 1.88 | -0.31 |
| MutationAssessor transf | low impact | low impact | medium impact |
| MutationAssessor transf score | -1.7 | -1.56 | -1 |
| CHASM | Neutral | Neutral | Neutral |
| CHASM pvalue | 0.83 | 0.53 | 0.28 |
| CHASM FDR | 0.9 | 0.8 | 0.8 |
| ClinVar id | 9710.0 | . | . |
| ClinVar Allele id | 24749.0 | . | . |
| ClinVar CLNDISDB | MONDO:MONDO:0010772,MedGen:C1839040,OMIM:500001,Orphanet:99718|Human_Phenotype_Ontology:HP:0001086,Human_Phenotype_Ontology:HP:0001112,MONDO:MONDO:0010788,MedGen:C0917796,OMIM:535000,Orphanet:104|MONDO:MONDO:0009723,MedGen:C0023264,OMIM:256000,Orphanet:506 | . | . |
| ClinVar CLNDN | Leber_optic_atrophy_and_dystonia|Leber_optic_atrophy|Leigh_syndrome | . | . |
| ClinVar CLNSIG | Benign | . | . |
| MITOMAP Disease Clinical info | LHON / LDYT / DEAF / hypertension helper mut. | . | . |
| MITOMAP Disease Status | Reported / possibly synergistic | . | . |
| MITOMAP Disease Hom/Het | +/+ | ./. | ./. |
| MITOMAP General GenBank Freq | 0.597% | . | . |
| MITOMAP General GenBank Seqs | 365 | . | . |
| MITOMAP General Curated refs | 21978175;18676632;17723226;18223312;21482521;19818876;35104579;24467713;8644732;27159682;15972314;29253894;16714301;24062162;35801081;20301353;17922426;17300996;18639500;29387390;24002810;29987491;17123466;16364244;21457906;22487888 | . | . |
| MITOMAP Variant Class | polymorphism;disease | . | . |
| gnomAD 3.1 AN | 56427.0 | . | . |
| gnomAD 3.1 AC Homo | 56.0 | . | . |
| gnomAD 3.1 AF Hom | 0.000992433 | . | . |
| gnomAD 3.1 AC Het | 0.0 | . | . |
| gnomAD 3.1 AF Het | 0.0 | . | . |
| gnomAD 3.1 filter | PASS | . | . |
| HelixMTdb AC Hom | 264.0 | . | . |
| HelixMTdb AF Hom | 0.0013470557 | . | . |
| HelixMTdb AC Het | 8.0 | . | . |
| HelixMTdb AF Het | 4.081987e-05 | . | . |
| HelixMTdb mean ARF | 0.35087 | . | . |
| HelixMTdb max ARF | 0.65169 | . | . |
| ToMMo 54KJPN AC | 553 | . | . |
| ToMMo 54KJPN AF | 0.010184 | . | . |
| ToMMo 54KJPN AN | 54302 | . | . |
| COSMIC 90 | . | . | . |
| dbSNP 156 id | rs200873900 | . | . |